RNAi is a naturally occuring process in which small RNA molecules activate a cellular process that results in the destruction of a specific mRNA.
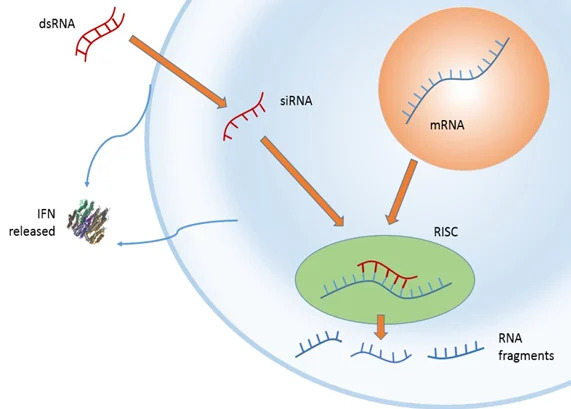
Image adapted from "RNAi Therapeutics: How Likely, How Soon?" Robinson R, PLos Biology, Vol 2, No. 1.e28 doi:10.1371/journal.pbio.0020028" under Creative Commons Attribution 2.5. Generic license.
RNA interference endogenous pathways
Endogenous RNAi pathways can be activated by double-stranded RNA of viruses (e.g. their replicative intermediate), transcripts from repetitive sequences in the host's genome, transposons, primary microRNA, and foreign DNA. The RNA polymerase II and III enzymes, Drosha and DICER, can cleave the double-stranded RNA and primary-miRNA (pri-RNA) and pre-miRNA into smaller pieces. Mature miRNA are usually 22 nt long. Double-stranded RNA of 30nt or longer usually induces interferon, which activates interferon response genes and proteins, a hallmark of the innate immune response.
Uses of shRNA, miRNA, siRNAs
Scientists use RNAi agents to probe gene function, such as genes involved in normal growth, maintenance, and repair processes; autoimmune diseases; viral replication; immune responses, and cancer.1-3
SiRNAs also are used to elucidate effector-mediator signal transduction pathways in normal cells, tumor cells, and chronic diseases 2,4,5 or elucidate genes modulated by the innate immune system in virally-infected cells or autoimmune diseases.5-7
SiRNA can help identify suitable targets for development of therapeutics,1,3,6 assess the mechanism of various medications,3,8 and be developed as novel therapeutics for pathogenic viruses, prions, and cancer.3,4,8-12
Specificity
RNAi strategies depend on its sequence-specific binding to mRNA, thus forming double-stranded RNA (dsRNA). A major benefit of this antisense technology is its theoretical specificity.
However, siRNA can induce gene silencing or modulation of various off-target mRNAs with less than 100% homology by matching in the 3’ untranslated region (UTR).13 The sense strand of the shRNA also causes off-target gene silencing.14
Since off-target mRNAs can differ between species,15 off-target effects of a specific siRNA, shRNA, or miRNA in human cells or a clinical trial can not be predicted by pre-clinical rodent models.16
3 Distinct triggers of off-target effects
The introduction of siRNA, shRNA, or dual/triple miRNA constructs into cells may induce non-specific side effects by three separate origins:
-
Lipid-mediated effects
-
RNA-induced silencing complex (RISC)-dependent off-target effects
-
Interferon response
SiRNA delivery with cationic lipids alters gene expression profiles of many genes, including interferon response genes.17 Some of these effects can be reduced by JAK inhibitors.17
Saturation of the RNA-induced silencing complex (RISC) with high quantities of shRNA driven by polymerase III transcription can induce off-target effects, such as cellular toxicity.18 Switching to polymerase II-driven transcription in the vector lowers shRNA production which reduces toxicity.18
Five receptors detect RNA and induce type I interferon production
Exogenous RNA can trigger receptors involved in four distinct signaling pathways which induce the production of type I interferon (IFN).11 Type I IFNs include IFN-α, IFN-β, IFN-ε, IFN-δ, IFN-τ, IFN-ζ, IFN-κ, IFN-ω, and IFN-ν. The four distinct pathways are:
-
Double-stand RNA (dsRNA)-dependent protein kinase (PKR) pathway activates in the presence of long (>33bp) ds RNA in a non-sequence-specific fashion.
-
Toll-like receptor-3 (TLR3)-induced pathway is stimulated by dsRNA bound to TLR3.
-
TLR7/ TLR8 –induced pathway is activated by synthetic nucleoside analogs, imidazoquinolines, and GU-rich short ssRNAs bound to the TLR7or TLR8 receptors.
-
Melanoma differentiation-associated protein 5 (MDA5)/ retinoic acid-inducible gene-I (RIG-I) pathway responds to dsRNAs, ssRNAs, siRNAs, and small-self RNAs.
-
MDA5 detects long RNA molecules that are not mRNA nor ribosomal RNA.
-
The RIG-I recognizes the nascent 5’-triphosphate moiety of viral transcripts; the negative-sense, single-stranded RNA (ssRNA) genome of viruses; short blunt-ended dsRNA of 21 nt long; RNase L-generated small-self RNAs; and dsRNAs of 300 to 1000 base pairs (bp).
Several miRNA can regulate IFN production, and IFN can regulate at least 36 distinct miRNA species.19
Type I Interferon can act as a detrimental off-target effect, an adjuvant, or a confounding factor. The siRNAs and the vectors may have intrinsic immunostimulatory activity11 that affects the inflammatory status of the targeted tissue.
The RNAimmuno database stores a searchable list of published data on the effects of various siRNAs, miRNAs, vectors, and reagents on the innate immune response in vitro and in vivo.20
Off-target effect:
Since interferon alpha can attenuate DICER-mediated processing of shRNA to siRNA but not siRNA-mediated knockdown of its target gene,21 shRNA-induced IFN production may decrease the amount of siRNA in the cells. The lower siRNA quantity in the cells may limit its gene silencing ability.
Interferon also reduces the efficacy of oncolytic Semliki Forest Virus for the treatment of glioblastoma in vitro and in mouse models.10
If siRNAs specific for overexpressed normal proteins in diseases also induce type I interferons, they can induce an inflammatory environment that may become detrimental. Various methods for reducing the siRNA-induced IFN production have been reported.11,22
Potential adjuvant:
SiRNAs and other antisense technology can reduce the replicative ability of prions,9 and viral genomes such as Hepatitis C virus and HIV.4,12,23,24
Confounding factor:
SiRNA-based exploration of gene function can be complicated by siRNA’s immunostimulatory effects. Thus, numerous laboratories are reporting modifications to reduce siRNA’s effect on innate immune responses, as recently reviewed.11 Although siRNA screening may identify many genes that are false-positive molecular targets, SiRNA controls that distinguish between on-target and off-target gene silencing have been reported.27
As aforementioned, siRNA regulates off-target transcripts in a species-specific manner.15 Thus, preclinical rodent models can not predict off-target effects of a specific siRNA, shRNA or miRNA in clinical trials.16
Thus, monitoring of off-target activity such as interferon production concurrent with siRNA experimentation and development may help clarify the interpretation of your results. Interferons (IFNα, IFNβ, and IFNγ) production can be efficiently and accurately monitored in plasma, culture supernatants, normal sera, and autoimmune serum 1,6,8,28,29 and CCL529 by ELISAs. The most commonly used assays are listed in Table 1. Purified IFNα or IFNβ from have been used as a standard in multiple assays for diverse species, including human,3 macaques,24 rodents (mouse, rat).31 For example, IFNα2 titrations are added as a standard in a functional interferon assay.32 A purified IFN added to a cellular assay can confirm the biological actions of interferon on distinct pathways.5,7,23,31
Table 1: Common ELISA assays for monitoring off-target Type I IFN production
Species
IFN
Sample Type
Comments
Product number
Human
Alpha (all subtypes)
Plasma, serum, TCM
Tolerates autoimmune serum
41115
Beta
Plasma, serum, TCM
High sensitive, Tolerates autoimmune serum, validated for IFN-Beta therapeutics
41415
Mouse
Alpha (all 14 subtypes)
Plasma, serum, TCM
High sensitive
42115
Beta
Plasma, serum, TCM
High sensitive
42410
References:
1. Smith N, Pietrancosta N, Davidson S, et al. Natural amines inhibit activation of human plasmacytoid dendritic cells through CXCR4 engagement. Nat Commun. 2017;8:14253.
2. Ueno N, Nishimura N, Ueno S, et al. PU.1 acts as tumor suppressor for myeloma cells through direct transcriptional repression of IRF4. Oncogene. 2017.
3. Beug ST, Beauregard CE, Healy C, et al. Smac mimetics synergize with immune checkpoint inhibitors to promote tumour immunity against glioblastoma. Nat Commun. 2017;8.
4. Usami Y, Wu Y, Gottlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526(7572):218-223.
5. Wang Q, Huang L, Hong Z, et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017;13(3):e1006264.
6. Dudhgaonkar S, Ranade S, Nagar J, et al. Selective IRAK4 Inhibition Attenuates Disease in Murine Lupus Models and Demonstrates Steroid Sparing Activity. J Immunol. 2017;198(3):1308-1319.
7. Li Y, Wu S, Pu J, Huang X, Zhang P. Dengue virus up-regulates expression of notch ligands Dll1 and Dll4 through interferon-beta signalling pathway. Immunology. 2015;144(1):127-138.
8. Kim SY, Noh YW, Kang TH, et al. Synthetic vaccine nanoparticles target to lymph node triggering enhanced innate and adaptive antitumor immunity. Biomaterials. 2017;130:56-66.
9. Kang SG, Kim C, Aiken J, Yoo HS, McKenzie D. Dual MicroRNA to Cellular Prion Protein Inhibits Propagation of Pathogenic Prion Protein in Cultured Cells. Mol Neurobiol. 2017.
10. Ramachandran M, Yu D, Dyczynski M, et al. Safe and Effective Treatment of Experimental Neuroblastoma and Glioblastoma Using Systemically Delivered Triple MicroRNA-Detargeted Oncolytic Semliki Forest Virus. Clin Cancer Res. 2017;23(6):1519-1530.
11. Meng Z, Lu M. RNA Interference-Induced Innate Immunity, Off-Target Effect, or Immune Adjuvant? Front Immunol. 2017;8:331.
12. Swamy MN, Wu H, Shankar P. Recent advances in RNAi-based strategies for therapy and prevention of HIV-1/AIDS. Adv Drug Deliv Rev. 2016;103:174-186.
13. Birmingham A, Anderson EM, Reynolds A, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3(3):199-204.
14. Mockenhaupt S, Grosse S, Rupp D, Bartenschlager R, Grimm D. Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc Natl Acad Sci U S A. 2015;112(30):E4007-4016.
15. Burchard J, Jackson AL, Malkov V, et al. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15(2):308-315.
16. Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57-67.
17. Tao W, Mao X, Davide JP, et al. Mechanistically probing lipid-siRNA nanoparticle-associated toxicities identifies Jak inhibitors effective in mitigating multifaceted toxic responses. Mol Ther. 2011;19(3):567-575.
18. Toro Cabrera G, Mueller C. Design of shRNA and miRNA for Delivery to the CNS. Methods Mol Biol. 2016;1382:67-80.
19. Forster SC, Tate MD, Hertzog PJ. MicroRNA as Type I Interferon-Regulated Transcripts and Modulators of the Innate Immune Response. Front Immunol. 2015;6:334.
20. Olejniczak M, Galka-Marciniak P, Polak K, Fligier A, Krzyzosiak WJ. RNAimmuno: a database of the nonspecific immunological effects of RNA interference and microRNA reagents. RNA. 2012;18(5):930-935.
21. Machitani M, Sakurai F, Wakabayashi K, Takayama K, Tachibana M, Mizuguchi H. Type I Interferons Impede Short Hairpin RNA-Mediated RNAi via Inhibition of Dicer-Mediated Processing to Small Interfering RNA. Mol Ther Nucleic Acids. 2017;6:173-182.
22. Sioud M. Overcoming the challenges of siRNA activation of innate immunity: design better therapeutic siRNAs. Methods Mol Biol. 2015;1218:301-319.
23. Metz P, Dazert E, Ruggieri A, et al. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012;56(6):2082-2093.
24. Shivakoti R, Hauer D, Adams RJ, et al. Limited in vivo production of type I or type III interferon after infection of macaques with vaccine or wild-type strains of measles virus. J Interferon Cytokine Res. 2015;35(4):292-301.
25. Zhang G, Chan B, Samarina N, et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A. 2016;113(8):E1034-1043.
26. Maine CJ, Teijaro JR, Marquardt K, Sherman LA. PTPN22 contributes to exhaustion of T lymphocytes during chronic viral infection. Proc Natl Acad Sci U S A. 2016;113(46):E7231-E7239.
27. Buehler E, Chen YC, Martin S. C911: A bench-level control for sequence specific siRNA off-target effects. PLoS One. 2012;7(12):e51942.
28. Zhan Z, Cao H, Xie X, et al. Phosphatase PP4 Negatively Regulates Type I IFN Production and Antiviral Innate Immunity by Dephosphorylating and Deactivating TBK1. J Immunol. 2015;195(8):3849-3857.
29. Imaizumi T, Hayakari R, Matsumiya T, et al. Chloroquine attenuates TLR3/IFN-beta signaling in cultured normal human mesangial cells: A possible protective effect against renal damage in lupus nephritis. Mod Rheumatol. 2017:1-18.
30. Bernardo AR, Cosgaya JM, Aranda A, Jimenez-Lara AM. Pro-apoptotic signaling induced by Retinoic acid and dsRNA is under the control of Interferon Regulatory Factor-3 in breast cancer cells. Apoptosis. 2017.
31. Mustachio LM, Lu Y, Tafe LJ, et al. Deubiquitinase USP18 Loss Mislocalizes and Destabilizes KRAS in Lung Cancer. Mol Cancer Res. 2017.
32. Jonsson KL, Laustsen A, Krapp C, et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat Commun. 2017;8:14391.
RNAi is a naturally occuring process in which small RNA molecules activate a cellular process that results in the destruction of a specific mRNA.

Image adapted from "RNAi Therapeutics: How Likely, How Soon?" Robinson R, PLos Biology, Vol 2, No. 1.e28 doi:10.1371/journal.pbio.0020028" under Creative Commons Attribution 2.5. Generic license.
RNA interference endogenous pathways
Endogenous RNAi pathways can be activated by double-stranded RNA of viruses (e.g. their replicative intermediate), transcripts from repetitive sequences in the host's genome, transposons, primary microRNA, and foreign DNA. The RNA polymerase II and III enzymes, Drosha and DICER, can cleave the double-stranded RNA and primary-miRNA (pri-RNA) and pre-miRNA into smaller pieces. Mature miRNA are usually 22 nt long. Double-stranded RNA of 30nt or longer usually induces interferon, which activates interferon response genes and proteins, a hallmark of the innate immune response.
Uses of shRNA, miRNA, siRNAs
Scientists use RNAi agents to probe gene function, such as genes involved in normal growth, maintenance, and repair processes; autoimmune diseases; viral replication; immune responses, and cancer.1-3
SiRNAs also are used to elucidate effector-mediator signal transduction pathways in normal cells, tumor cells, and chronic diseases 2,4,5 or elucidate genes modulated by the innate immune system in virally-infected cells or autoimmune diseases.5-7
SiRNA can help identify suitable targets for development of therapeutics,1,3,6 assess the mechanism of various medications,3,8 and be developed as novel therapeutics for pathogenic viruses, prions, and cancer.3,4,8-12
Specificity
RNAi strategies depend on its sequence-specific binding to mRNA, thus forming double-stranded RNA (dsRNA). A major benefit of this antisense technology is its theoretical specificity.
However, siRNA can induce gene silencing or modulation of various off-target mRNAs with less than 100% homology by matching in the 3’ untranslated region (UTR).13 The sense strand of the shRNA also causes off-target gene silencing.14
Since off-target mRNAs can differ between species,15 off-target effects of a specific siRNA, shRNA, or miRNA in human cells or a clinical trial can not be predicted by pre-clinical rodent models.16
3 Distinct triggers of off-target effects
The introduction of siRNA, shRNA, or dual/triple miRNA constructs into cells may induce non-specific side effects by three separate origins:
-
Lipid-mediated effects
-
RNA-induced silencing complex (RISC)-dependent off-target effects
-
Interferon response
SiRNA delivery with cationic lipids alters gene expression profiles of many genes, including interferon response genes.17 Some of these effects can be reduced by JAK inhibitors.17
Saturation of the RNA-induced silencing complex (RISC) with high quantities of shRNA driven by polymerase III transcription can induce off-target effects, such as cellular toxicity.18 Switching to polymerase II-driven transcription in the vector lowers shRNA production which reduces toxicity.18
Five receptors detect RNA and induce type I interferon production
Exogenous RNA can trigger receptors involved in four distinct signaling pathways which induce the production of type I interferon (IFN).11 Type I IFNs include IFN-α, IFN-β, IFN-ε, IFN-δ, IFN-τ, IFN-ζ, IFN-κ, IFN-ω, and IFN-ν. The four distinct pathways are:
-
Double-stand RNA (dsRNA)-dependent protein kinase (PKR) pathway activates in the presence of long (>33bp) ds RNA in a non-sequence-specific fashion.
-
Toll-like receptor-3 (TLR3)-induced pathway is stimulated by dsRNA bound to TLR3.
-
TLR7/ TLR8 –induced pathway is activated by synthetic nucleoside analogs, imidazoquinolines, and GU-rich short ssRNAs bound to the TLR7or TLR8 receptors.
-
Melanoma differentiation-associated protein 5 (MDA5)/ retinoic acid-inducible gene-I (RIG-I) pathway responds to dsRNAs, ssRNAs, siRNAs, and small-self RNAs.
-
MDA5 detects long RNA molecules that are not mRNA nor ribosomal RNA.
-
The RIG-I recognizes the nascent 5’-triphosphate moiety of viral transcripts; the negative-sense, single-stranded RNA (ssRNA) genome of viruses; short blunt-ended dsRNA of 21 nt long; RNase L-generated small-self RNAs; and dsRNAs of 300 to 1000 base pairs (bp).
-
Several miRNA can regulate IFN production, and IFN can regulate at least 36 distinct miRNA species.19
Type I Interferon can act as a detrimental off-target effect, an adjuvant, or a confounding factor. The siRNAs and the vectors may have intrinsic immunostimulatory activity11 that affects the inflammatory status of the targeted tissue.
The RNAimmuno database stores a searchable list of published data on the effects of various siRNAs, miRNAs, vectors, and reagents on the innate immune response in vitro and in vivo.20
Off-target effect:
Since interferon alpha can attenuate DICER-mediated processing of shRNA to siRNA but not siRNA-mediated knockdown of its target gene,21 shRNA-induced IFN production may decrease the amount of siRNA in the cells. The lower siRNA quantity in the cells may limit its gene silencing ability.
Interferon also reduces the efficacy of oncolytic Semliki Forest Virus for the treatment of glioblastoma in vitro and in mouse models.10
If siRNAs specific for overexpressed normal proteins in diseases also induce type I interferons, they can induce an inflammatory environment that may become detrimental. Various methods for reducing the siRNA-induced IFN production have been reported.11,22
Potential adjuvant:
SiRNAs and other antisense technology can reduce the replicative ability of prions,9 and viral genomes such as Hepatitis C virus and HIV.4,12,23,24
Confounding factor:
SiRNA-based exploration of gene function can be complicated by siRNA’s immunostimulatory effects. Thus, numerous laboratories are reporting modifications to reduce siRNA’s effect on innate immune responses, as recently reviewed.11 Although siRNA screening may identify many genes that are false-positive molecular targets, SiRNA controls that distinguish between on-target and off-target gene silencing have been reported.27
As aforementioned, siRNA regulates off-target transcripts in a species-specific manner.15 Thus, preclinical rodent models can not predict off-target effects of a specific siRNA, shRNA or miRNA in clinical trials.16
Thus, monitoring of off-target activity such as interferon production concurrent with siRNA experimentation and development may help clarify the interpretation of your results. Interferons (IFNα, IFNβ, and IFNγ) production can be efficiently and accurately monitored in plasma, culture supernatants, normal sera, and autoimmune serum 1,6,8,28,29 and CCL529 by ELISAs. The most commonly used assays are listed in Table 1. Purified IFNα or IFNβ from have been used as a standard in multiple assays for diverse species, including human,3 macaques,24 rodents (mouse, rat).31 For example, IFNα2 titrations are added as a standard in a functional interferon assay.32 A purified IFN added to a cellular assay can confirm the biological actions of interferon on distinct pathways.5,7,23,31
Table 1: Common ELISA assays for monitoring off-target Type I IFN production
| Species | IFN | Sample Type | Comments | Product number |
| Human | Alpha (all subtypes) | Plasma, serum, TCM | Tolerates autoimmune serum | 41115 |
| Beta | Plasma, serum, TCM | High sensitive, Tolerates autoimmune serum, validated for IFN-Beta therapeutics | 41415 | |
| Mouse | Alpha (all 14 subtypes) | Plasma, serum, TCM | High sensitive | 42115 |
| Beta | Plasma, serum, TCM | High sensitive | 42410 |
References:
1. Smith N, Pietrancosta N, Davidson S, et al. Natural amines inhibit activation of human plasmacytoid dendritic cells through CXCR4 engagement. Nat Commun. 2017;8:14253.
2. Ueno N, Nishimura N, Ueno S, et al. PU.1 acts as tumor suppressor for myeloma cells through direct transcriptional repression of IRF4. Oncogene. 2017.
3. Beug ST, Beauregard CE, Healy C, et al. Smac mimetics synergize with immune checkpoint inhibitors to promote tumour immunity against glioblastoma. Nat Commun. 2017;8.
4. Usami Y, Wu Y, Gottlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526(7572):218-223.
5. Wang Q, Huang L, Hong Z, et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017;13(3):e1006264.
6. Dudhgaonkar S, Ranade S, Nagar J, et al. Selective IRAK4 Inhibition Attenuates Disease in Murine Lupus Models and Demonstrates Steroid Sparing Activity. J Immunol. 2017;198(3):1308-1319.
7. Li Y, Wu S, Pu J, Huang X, Zhang P. Dengue virus up-regulates expression of notch ligands Dll1 and Dll4 through interferon-beta signalling pathway. Immunology. 2015;144(1):127-138.
8. Kim SY, Noh YW, Kang TH, et al. Synthetic vaccine nanoparticles target to lymph node triggering enhanced innate and adaptive antitumor immunity. Biomaterials. 2017;130:56-66.
9. Kang SG, Kim C, Aiken J, Yoo HS, McKenzie D. Dual MicroRNA to Cellular Prion Protein Inhibits Propagation of Pathogenic Prion Protein in Cultured Cells. Mol Neurobiol. 2017.
10. Ramachandran M, Yu D, Dyczynski M, et al. Safe and Effective Treatment of Experimental Neuroblastoma and Glioblastoma Using Systemically Delivered Triple MicroRNA-Detargeted Oncolytic Semliki Forest Virus. Clin Cancer Res. 2017;23(6):1519-1530.
11. Meng Z, Lu M. RNA Interference-Induced Innate Immunity, Off-Target Effect, or Immune Adjuvant? Front Immunol. 2017;8:331.
12. Swamy MN, Wu H, Shankar P. Recent advances in RNAi-based strategies for therapy and prevention of HIV-1/AIDS. Adv Drug Deliv Rev. 2016;103:174-186.
13. Birmingham A, Anderson EM, Reynolds A, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3(3):199-204.
14. Mockenhaupt S, Grosse S, Rupp D, Bartenschlager R, Grimm D. Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc Natl Acad Sci U S A. 2015;112(30):E4007-4016.
15. Burchard J, Jackson AL, Malkov V, et al. MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA. 2009;15(2):308-315.
16. Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57-67.
17. Tao W, Mao X, Davide JP, et al. Mechanistically probing lipid-siRNA nanoparticle-associated toxicities identifies Jak inhibitors effective in mitigating multifaceted toxic responses. Mol Ther. 2011;19(3):567-575.
18. Toro Cabrera G, Mueller C. Design of shRNA and miRNA for Delivery to the CNS. Methods Mol Biol. 2016;1382:67-80.
19. Forster SC, Tate MD, Hertzog PJ. MicroRNA as Type I Interferon-Regulated Transcripts and Modulators of the Innate Immune Response. Front Immunol. 2015;6:334.
20. Olejniczak M, Galka-Marciniak P, Polak K, Fligier A, Krzyzosiak WJ. RNAimmuno: a database of the nonspecific immunological effects of RNA interference and microRNA reagents. RNA. 2012;18(5):930-935.
21. Machitani M, Sakurai F, Wakabayashi K, Takayama K, Tachibana M, Mizuguchi H. Type I Interferons Impede Short Hairpin RNA-Mediated RNAi via Inhibition of Dicer-Mediated Processing to Small Interfering RNA. Mol Ther Nucleic Acids. 2017;6:173-182.
22. Sioud M. Overcoming the challenges of siRNA activation of innate immunity: design better therapeutic siRNAs. Methods Mol Biol. 2015;1218:301-319.
23. Metz P, Dazert E, Ruggieri A, et al. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology. 2012;56(6):2082-2093.
24. Shivakoti R, Hauer D, Adams RJ, et al. Limited in vivo production of type I or type III interferon after infection of macaques with vaccine or wild-type strains of measles virus. J Interferon Cytokine Res. 2015;35(4):292-301.
25. Zhang G, Chan B, Samarina N, et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc Natl Acad Sci U S A. 2016;113(8):E1034-1043.
26. Maine CJ, Teijaro JR, Marquardt K, Sherman LA. PTPN22 contributes to exhaustion of T lymphocytes during chronic viral infection. Proc Natl Acad Sci U S A. 2016;113(46):E7231-E7239.
27. Buehler E, Chen YC, Martin S. C911: A bench-level control for sequence specific siRNA off-target effects. PLoS One. 2012;7(12):e51942.
28. Zhan Z, Cao H, Xie X, et al. Phosphatase PP4 Negatively Regulates Type I IFN Production and Antiviral Innate Immunity by Dephosphorylating and Deactivating TBK1. J Immunol. 2015;195(8):3849-3857.
29. Imaizumi T, Hayakari R, Matsumiya T, et al. Chloroquine attenuates TLR3/IFN-beta signaling in cultured normal human mesangial cells: A possible protective effect against renal damage in lupus nephritis. Mod Rheumatol. 2017:1-18.
30. Bernardo AR, Cosgaya JM, Aranda A, Jimenez-Lara AM. Pro-apoptotic signaling induced by Retinoic acid and dsRNA is under the control of Interferon Regulatory Factor-3 in breast cancer cells. Apoptosis. 2017.
31. Mustachio LM, Lu Y, Tafe LJ, et al. Deubiquitinase USP18 Loss Mislocalizes and Destabilizes KRAS in Lung Cancer. Mol Cancer Res. 2017.
32. Jonsson KL, Laustsen A, Krapp C, et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat Commun. 2017;8:14391.